 |
|
|
Which DNA polymerase do Z-PCR Mix, 5x Z-PCR Mix, Direct-Load PCR Mix and 5xDirect-Load PCR Mix contain?
Z-PCR Mix, 5xZ-PCR Mix, Direct-Load PCR Mix and 5x Direct-Load PCR Mix contain TaqDNA polymerase.
Return to Top
What is Z-PCR Mix?
Z-PCR Mix is a complete ready-to-use reaction mixture optimized for efficient amplification of DNAs (genomic, plasmid, phage, or viral DNA) by PCR. Users only need to add
template and primers to set up a PCR reaction. Z-PCR Mix is designed to save time for setting up PCR reactions and reduce the risk of contamination.
Return to Top
What is 5xZ-PCR Mix?
5xZ-PCR Mix is a 5x ready-to-use reaction mixture optimized for efficient amplification of DNA (genomic, plasmid, phage, or viral DNA) by PCR. Users only need to add
a template, water and primers to set up a PCR reaction. 5xZ-PCR Mix contains stabilizers and loading buffer without dye, so that PCR products can be loaded directly onto an agarose gel after PCR. It is designed to save time when setting up PCR reactions and reduce the risk of contamination. The 5xZ-PCR Mix has the following advantages: a very diluted template DNA can be used in a PCR amplification and
5xZ-PCR Mix is more stable than 2x and 1.1x PCR Master Mixes.
Return to Top
What is Direct-Load PCR Mix?
Direct-Load PCR Mix contains all the components including loading dye (bromophenol blue) for PCR with the exception
of a template and primers. The advantage of Direct-Load PCR Mix is that samples can be loaded directly onto an agarose gel after PCR.
Return to Top
What is 5xDirect-Load PCR Mix?
5xDirect-Load PCR Mix is a newly formulated PCR Master Mix that improves the stability of enzymes and other components.
The 5x Direct-Load PCR Master Mix is an optimized ready-to-use 5x mixture containing everything (except a DNA template and
primer set) needed for efficient amplification of a template DNA (up to 3.5 kb) in PCR. The 5x Direct-Load PCR Master Mix is
designed to save time when setting up reactions, improve stability and reduce the risk of contamination. The 5x Direct-Load
PCR Master Mix contains stabilizers and loading dye (bromophenol blue) so that PCR products can be directly loaded onto agarose or polyacrylamide gels. PCR products can also
be digested with enzymes or used for ligation without prior removal of the loading dye. The 5x Direct-Load PCR Master Mix
is more stable than 2x and 1.1x PCR Master Mixes.
Return to Top
What is Z-PCR Mix GC-Rich?
Z-PCR Mix GC-Rich is a complete ready-to-use reaction mixture specifically optimized for efficient amplification of GC-rich and/or problematic
templates by PCR. Z-PCR Mix GC-Rich contains enhancing agents and stabilizers that can lower the Tm of the template. Z-PCR Mix GC-Rich
has been successfully tested for specific amplification of a 72.7% GC-rich region in human genomic DNA. Z-PCR Mix GC-Rich is designed to save time
when troubleshooting, improve stability and reduce the risk of contamination.
Return to Top
How does Z-PCR Mix GC-Rich compare with PCR Master Mixes from other companies for the amplification of GC-rich templates?
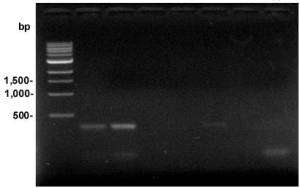
Z-PCR Mix GC-Rich has been compared with PCR Master Mixes from other companies for amplification of a 72.7% GC-rich region (348bp)
in human genomic DNA. Results show that three out of four PCR Master Mixes from other companies did not yield a specific DNA band (Figure 1,
Samples 3, 4, and 6). Z-PCR Mix GC-Rich (Sample 2) gives a much higher yield than the PCR Master Mix from
Company A (Sample 5) under the same conditions.
|
|
Z-BioMed
Other Companies
|
|
M 1
2 3
4 5 6 (-)
|
 |
| 1 Direct-Load PCR Mix GC-Rich Z-BioMed |
| 2 Z-PCR Mix
GC-Rich Z-BioMed |
| 3 PCR Master
Mix
Company G |
| 4 PCR Master
Mix
Company P |
| 5 PCR Master
Mix Company
A |
| 6 PCR Master
Mix Company
B |
|
Return to Top |
| Source |
Base pairs/ haploid genome |
Molecules per microgram (µg) |
Molecules per nanogram (ng) |
| 1 kb dsDNA |
_ |
9.12x 1011 |
9.12x 108 |
| 1 kb ssDNA |
_ |
1.82x 1012 |
1.82x 109 |
| 1 kb RNA |
_ |
1.77x 1012 |
1.77x 109 |
| Lambda DNA |
48,502 |
1.90x 1010 |
1.90x 107 |
|
E.coli genomic DNA
|
4.7x 106
|
1.94x 108
|
1.94x 105
|
|
S. cerevisiae genomic DNA
|
1.21x 107 |
7.54x 107
|
7.54x 104
|
|
C. elegans genomic DNA |
9.7x 107
|
9.40x 106
|
9.40x 103
|
|
A. thaliana
|
1.0x 108
|
9.12x 106
|
9.12x 103
|
|
Drosophila genomic DNA
|
1.8x 108
|
5.07x 106
|
5.07x 103
|
|
Mouse genomic DNA
|
3.0x 109
|
3.04x 105
|
3.04x 102
|
|
Rat genomic DNA
|
3.0x 109
|
3.04x 105
|
3.04x 102
|
|
Human genomic DNA
|
3.0x 109
|
3.04x 105
|
3.04x 102
|
Return to Top
What concentration of primers should I use for my PCR?
0.1 µM to 1.5 µM of each primer should be used for a PCR.
Return to Top
How to convert pmoles of primers to µM of primers?
1 pmole/µl = 1 µM; 1 nmole/µl = 1 mM
For example, if you have 60500 pmoles (60.5 nmoles) of primer, you can add 60.5 µl of
dH20 to make a 1mM stock solution of primer (60.5 nmoles + 60.5 µl of dH20 = 1 mM solution of primer).
Dilute 1:100 = 10 µM
If you use 1 µl of 10 µM primer in 25 µl PCR reaction, your final primer concentration will be 0.5 µM.
Return to Top
When should I use a Taq DNA polymerase?
If you want to amplify DNA fragments efficiently, you should use a Taq DNA polymerase. Z-PCR Mix, Direct-Load PCR Mix and
Z-PCR Mix GC-Rich contain Taq DNA polymerase at the optimized
concentration which can give a high efficiency and yield. Nonproofreading enzymes such as Taq DNA polymerase can give you minimum errors and maximum efficiency under appropriate high-fidelity conditions. High-fidelity conditions
include low magnesium and dNTP concentrations (<50µM), and short extension times. It is very hard to beat Taq DNA polymerase for the combination of specificity, sensitivity, and
yield.
Return to Top
When should I use proofreading enzymes such as Pfu, Pwo, Vent or Deep Vent?
If you need a short fragment from a plasmid, use a high template ( i.e ., ng of plasmid) and keep the cycle number low (10 or 15), then it is fine to
use proofreading DNA polymerases. However, you can not assume that the PCR product will have 100% fidelity just by using a proofreading DNA
polymerase. No DNA polymerase is perfect. That is, when fidelity is important you still have to sequence your clone no matter how
accurate the proofreading DNA polymerase is.
Return to Top
How does Taq DNA polymerase compare with proofreading enzymes such as Pfu, Vent and Deep Vent?
The properties of Taq and other proofreading DNA polymerases are listed in Table 2.
|
| Table 2. Comparison of Taq and Some Proofreading DNA polymerases |
| DNA Polymerase |
Taq
|
Pfu
|
Vent
|
Deep Vent
|
|
Organism
|
Thermus Aquaticus
|
Pyrococcus Furiosus
|
Thermococcus Litoralis
|
Pyrococcus Species GB-1
|
|
5'-3' Exonuclease Activity
|
Yes
|
No
|
No
|
No
|
|
3'-5' Exonuclease Activity
|
No
|
Yes
|
Yes
|
Yes
|
|
PCR DNA Ends
|
3'-A
|
Blunt
|
95% Blunt
|
95% Blunt
|
|
Yield
|
+ + + +
|
+ +
|
+ +
|
+ +
|
|
Error Rate (error/bp incorporated)
|
8 x 10-6
|
1.3 x 10-6
|
2.8 x 10-6
|
2.7 x 10-6
|
PCR Mutation Calculator
To calculate the percentage of PCR-induced mutants with different polymerases such as Taq, Pfu, Vent and Deep Vent in any PCR products, enter the target size and cycling number to the following table.
|



